The excellence in One Health
Nano1Health SL is a company based on rapidly and accurately identifying microorganisms and pathogens from a One Health perspective.
Our B2B company boasts a diverse team combining the unparalleled expertise of veterinarians, geneticists, and bioinformaticians, who are leaders in their respective fields and widely recognized for their significant contributions and achievements.
What we do?
We develop innovative strategies for rapidly identifying microorganisms, pathogens, and drug-resistance biomarkers. Our solutions are based on nanopore genomic sequencing and expert bioinformatics analysis.
We offer in-depth characterization of bacteria, fungi, and protozoa through whole-genome sequencing (WGS) using nanopore technology from microbiological cultures. Our analysis includes genome assembly, gene annotation, and functional characterization. Additionally, we provide SNP and structural variant analysis.
We identify antibiotic and drug resistance biomarkers by leveraging similarity-based database searches and predictive models. This work is crucial in informing therapeutic strategies for clinical and industrial applications, contributing to the global effort in combating drug resistance.
- Antibiotic resistance in bacteria (AMR)
-
Drug resistance in Leishmania infantum
-
Cultures (>5*10E7 p/ml) - LeishGenApp
-
Clinical samples (>5.000 p/ml) - LeishGenR
-
Our agnostic metagenomic sequencing process is designed for rapid pathogen identification. It efficiently detects viruses, bacteria, fungi, and parasites, providing quick results.
-
PIP (Pathogen Identification Platform) utilizes metagenomic algorithms and a proprietary database to generate a list of potential pathogens from complex clinical samples.
Our comprehensive analysis of the full-length 16S rRNA gene (>1.4 kb) ensures precise genus and species identification of complex microbial communities across various sample types. With a proprietary database covering over 25,000 distinct species, it provides thoroughness of the microbiome profiling.
We offer unbiased, agnostic identification and classification of microorganisms (bacteria, viruses, fungi, and parasites) from complex samples, revealing their functional potential and taxonomic diversity.
Our cloud-based bioinformatics platform provides user-friendly analysis of nanopore sequencing data from microorganisms. It is designed to support professionals in basic research, clinical applications, and industrial portfolios and offers long-term data storage.
As a Contract Research Organization (CRO), we provide expert consulting services tailored to our clients' projects. Our commitment to client satisfaction is evident in our support for the entire data generation process, from sequencing to the design and customization of bioinformatics algorithms and pipelines based on specific research needs.
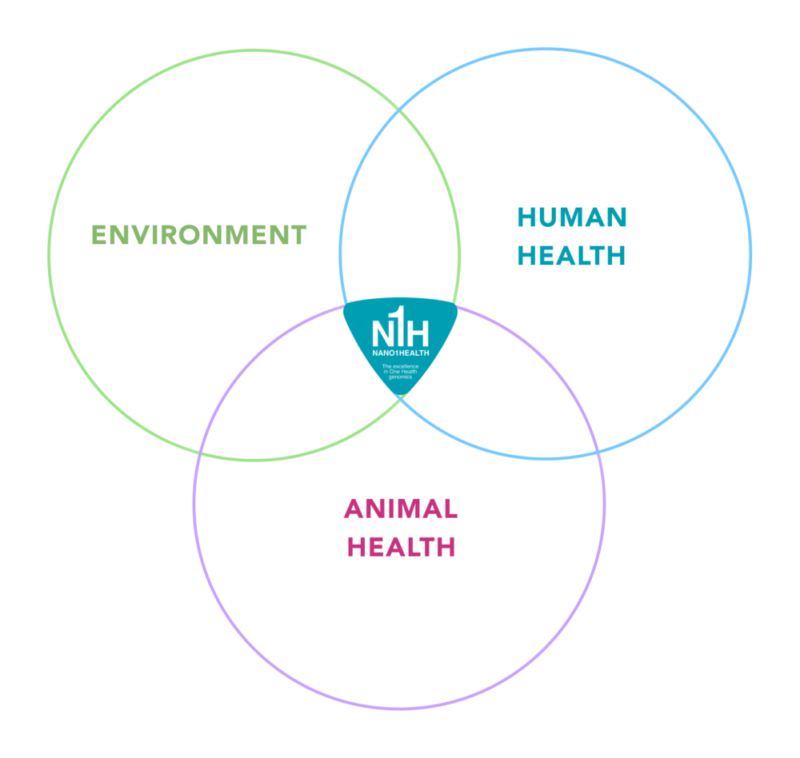
How?
Our multidisciplinary team stands out for its long history and experience in genomics, bioinformatics, and veterinary clinics. We offer a close and personalized contact for each client.

Knowledge
The team's previous experience and complementarity.

Innovation
State-of-the-art tools, ready to offer innovative, accurate, and affordable solutions.

Unique
With a One Health vision
Who?
We are a multidisciplinary team that combines the experience of veterinarians, geneticists, and bioinformaticians of recognized prestige.
Our clients
We work in different sectors with extensive research backgrounds.
- Pharmaceutical, veterinary, biomedical, biomedical, and agri-food research professionals.
- Research groups or companies with R+D+i projects
- Professionals in the agri-food sector
- Public and private research professionals